Hello,
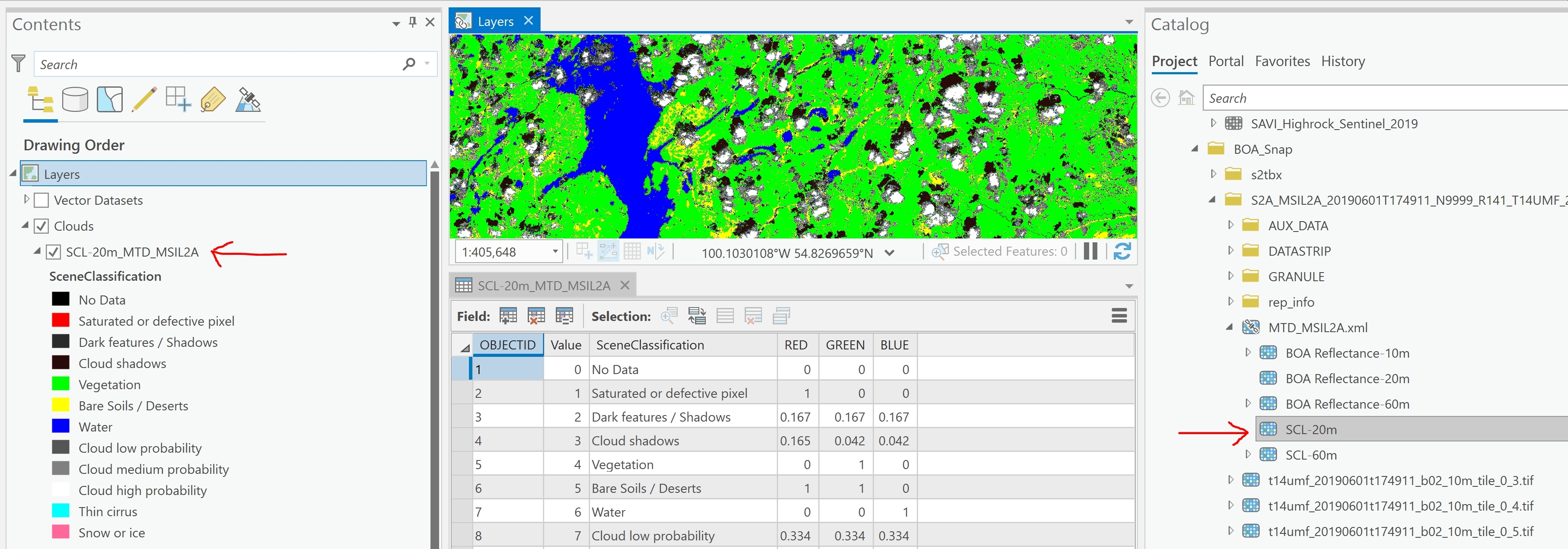
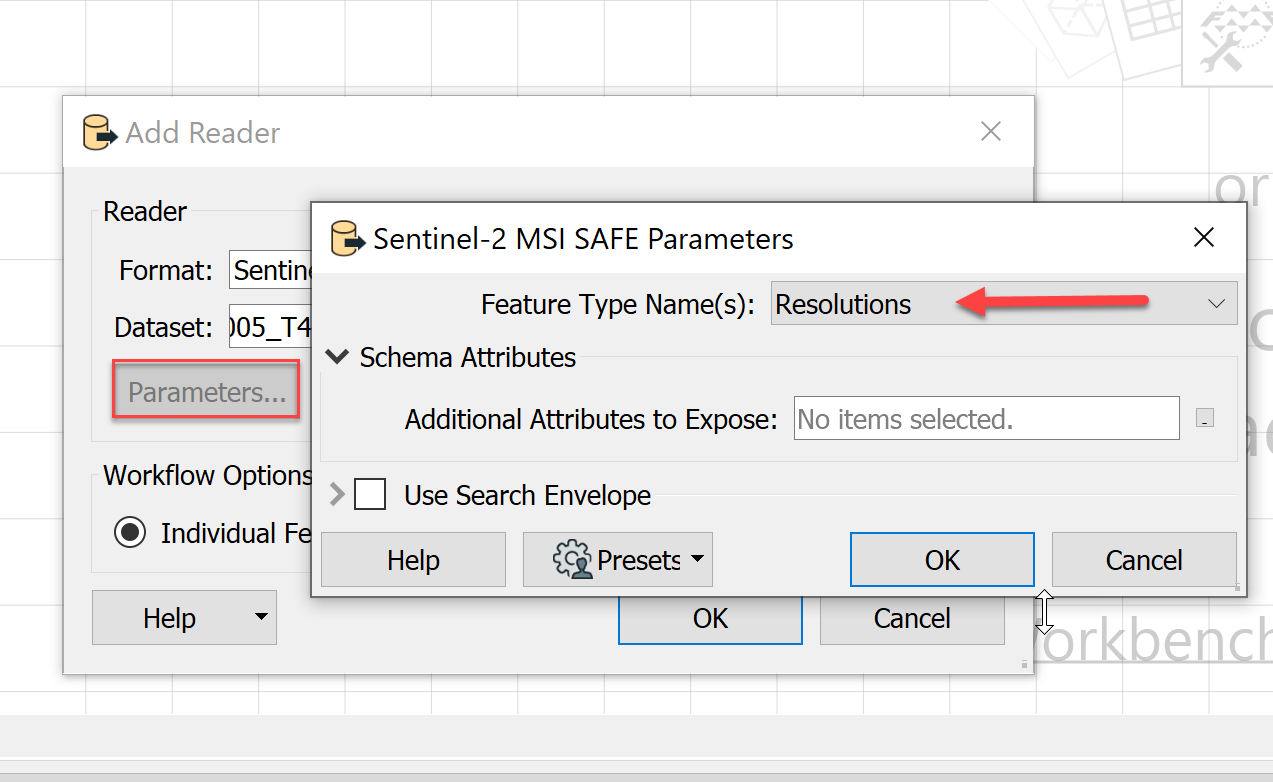
I've converted Sentinel-2 Level-1C scenes into Level-2A bottom of atmosphere (BOA) products using the ESA SNAP Sentinel Toolbox. I would now like to use FME to read the scene classification (SCL-20m) SNAP outputs and create a raster mosaic of these. The attached illustration shows this data in ArcGIS Pro. I've been unable to read the data in FME.
Thanks.
Tony