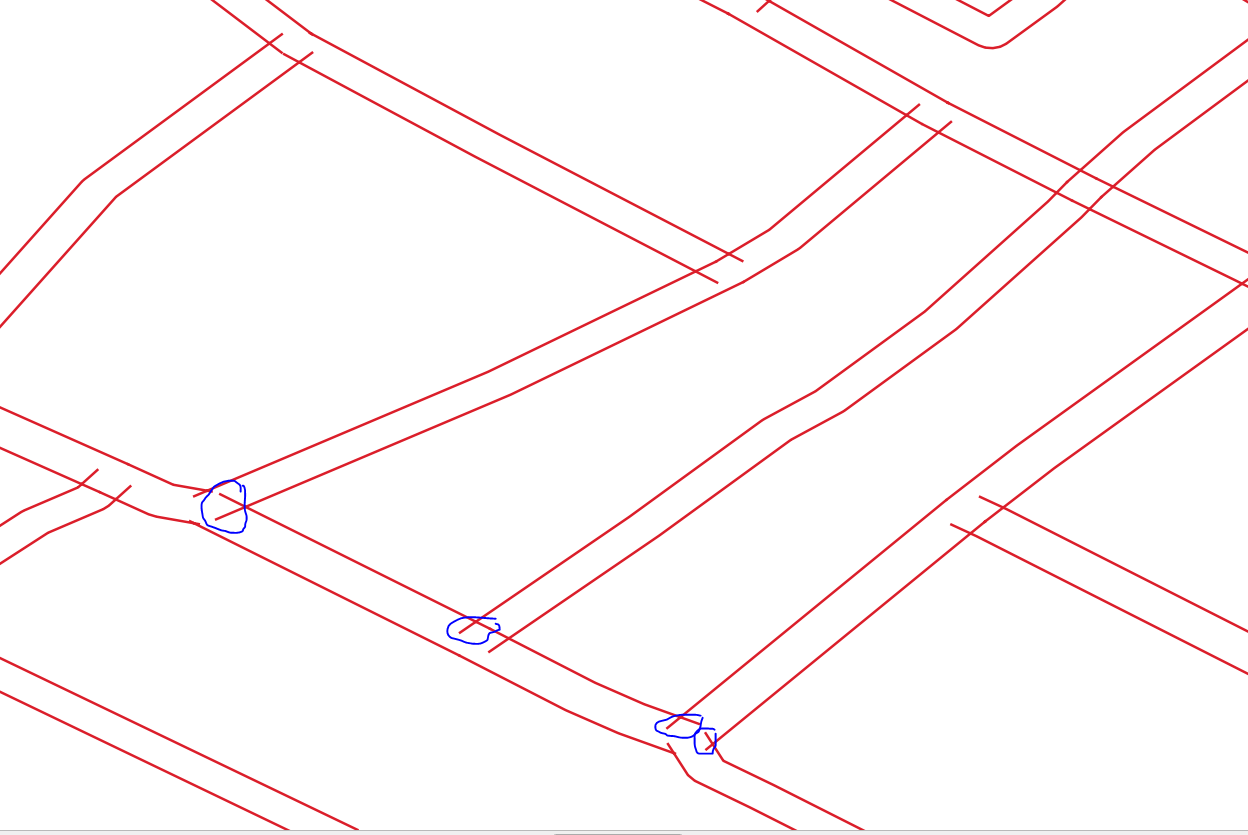
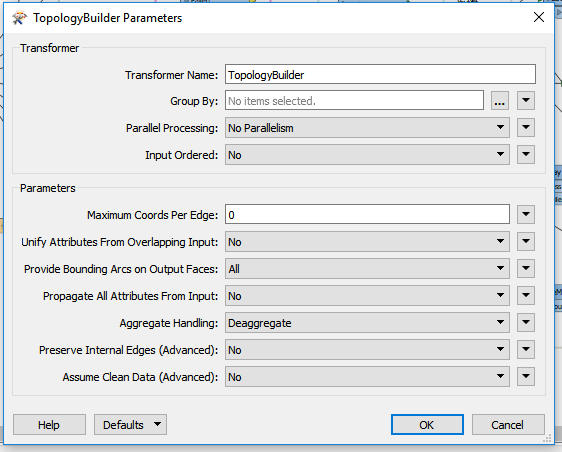
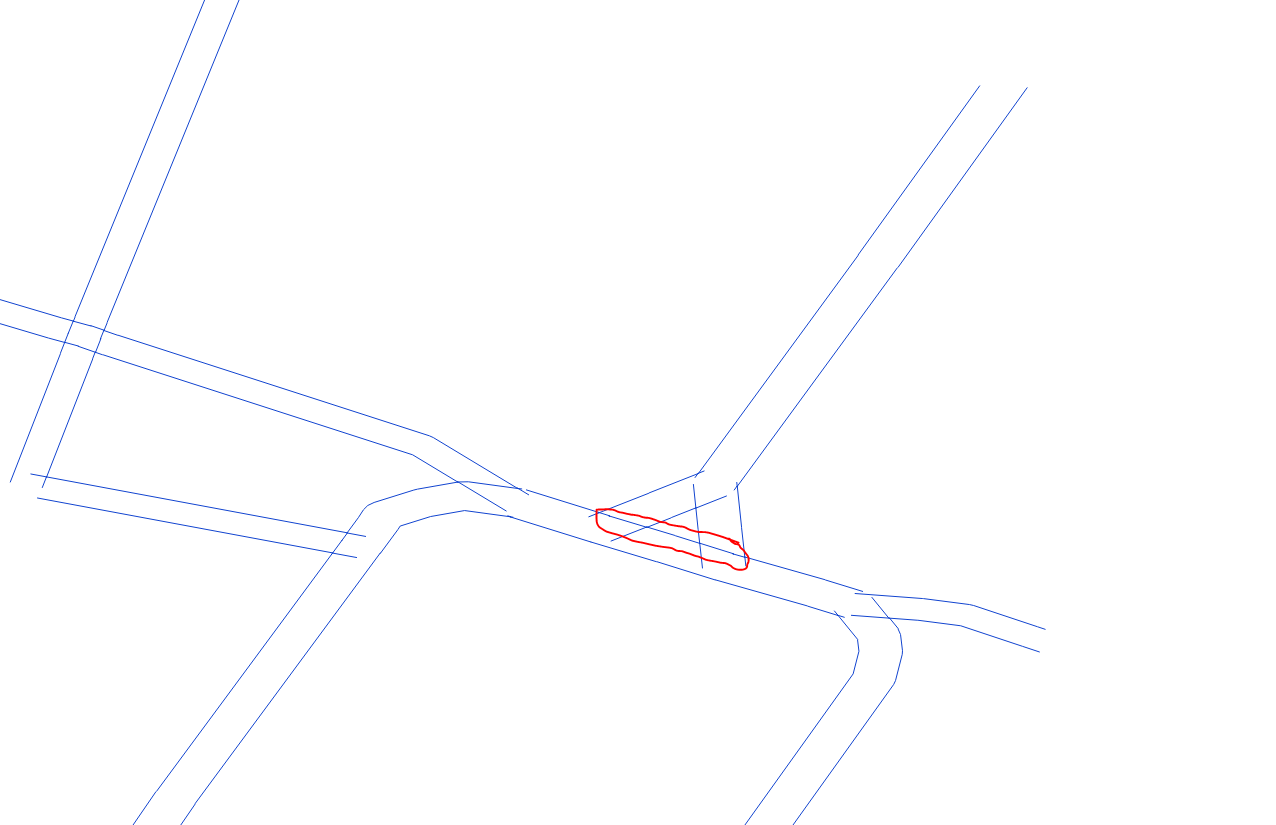
@jugoslaviaa Thanks for including some sample data. The problem here is that the data is very poor quality, with lines that overlap and lines that have gaps. So it's very unlikely that you'll get a result that removes 100% of the dangles, without also removing valid edges - see the purple edges in the image below). As has been suggested, TopologyBuilder is your friend, but then you can post process the TopologyBuilder output to help identify the valid/invalid edges, or edges that are ambiguous.
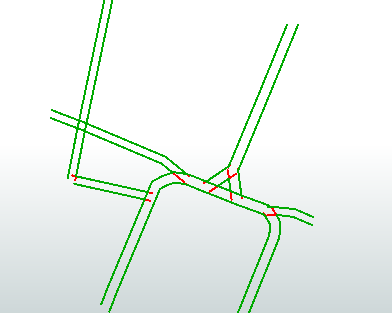
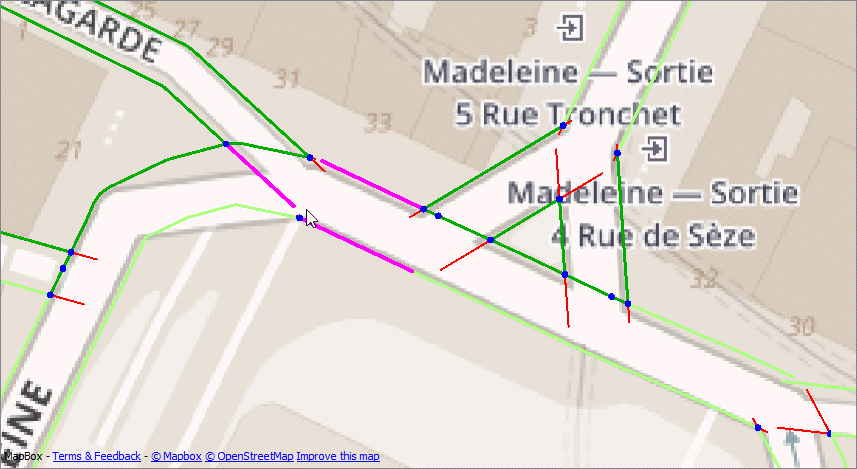
The attached workspace finds the nodes that touch more than one edge - internal nodes (Blue). The end of line nodes are dropped. Then we can test for:
- any edge that has internal nodes at either end is an internal edge and should be kept (dark green)
- any edge longer than x (I've used 20m) should be kept (light green)
- any edge that is short (>10m) and only has one node is probably a dangle - reject (red)
- any edge that has one internal node and is between 10 & 20 m is ambiguous and should be validated manually (purple)
You'll probably find other things you can test for when you work with a larger dataset
Workspace:
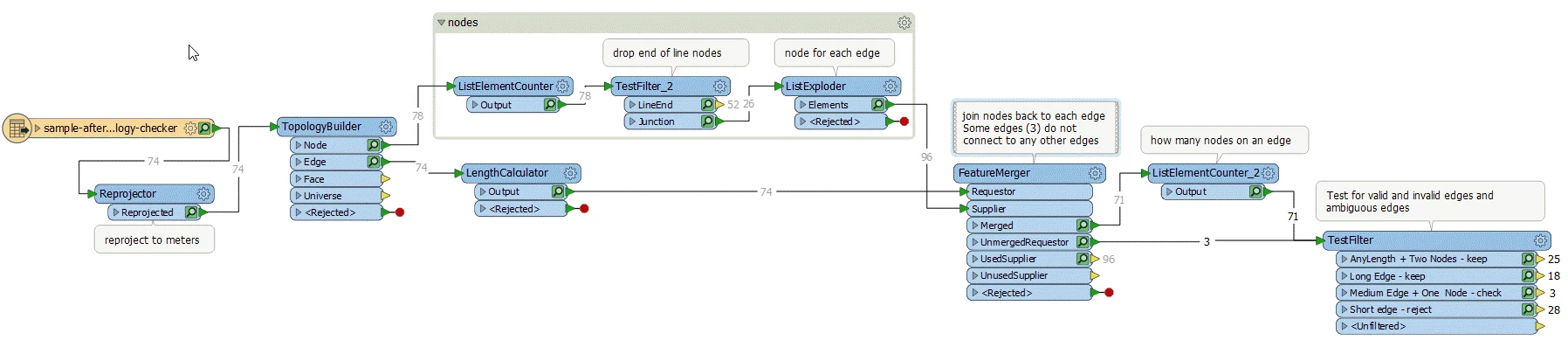
Workspace (2018.1):finddangles.fmw