I want to cut the lines inside a polygon. I thought of using the Clipper transformer however it is very slow. any ideas how I can optimize it and make it faster.
see below:
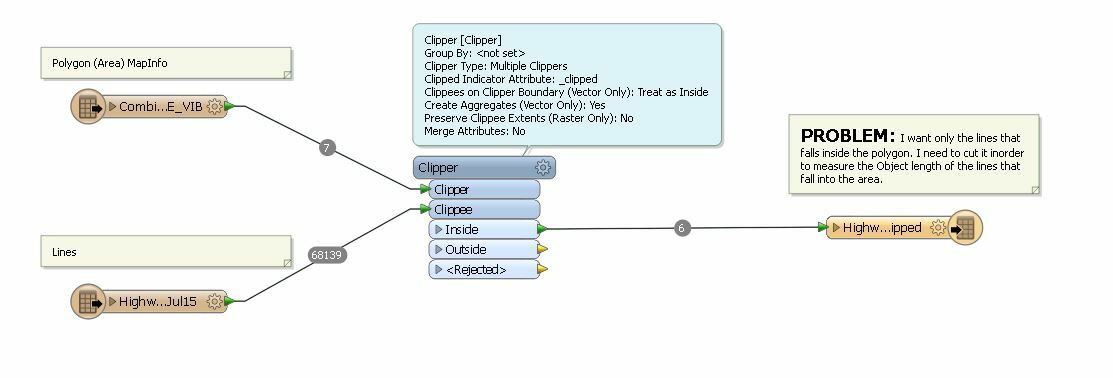
been a few mins now and looks like it only finished 6 lines..leaving it running until I think of another way to do it.


















