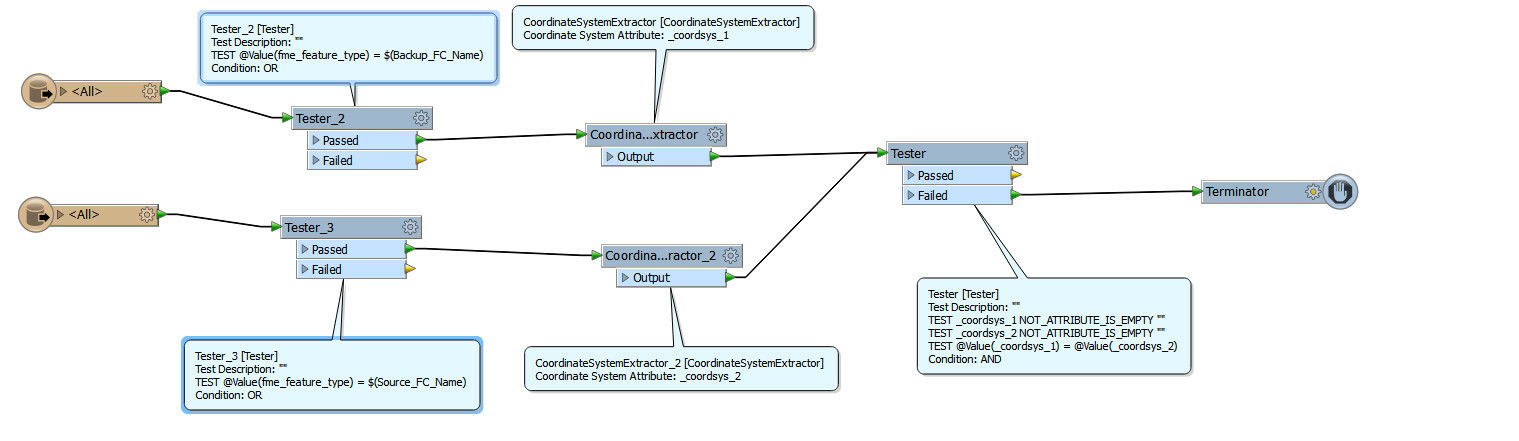
I am trying to compare corrdinate system between 2 feature class . If corrdinate system is different then I want to terminate the process
In this below workflow my tester always gets failed even when both feature class are in same coordinate system .
Can anyoone point out what am I doing wrong .