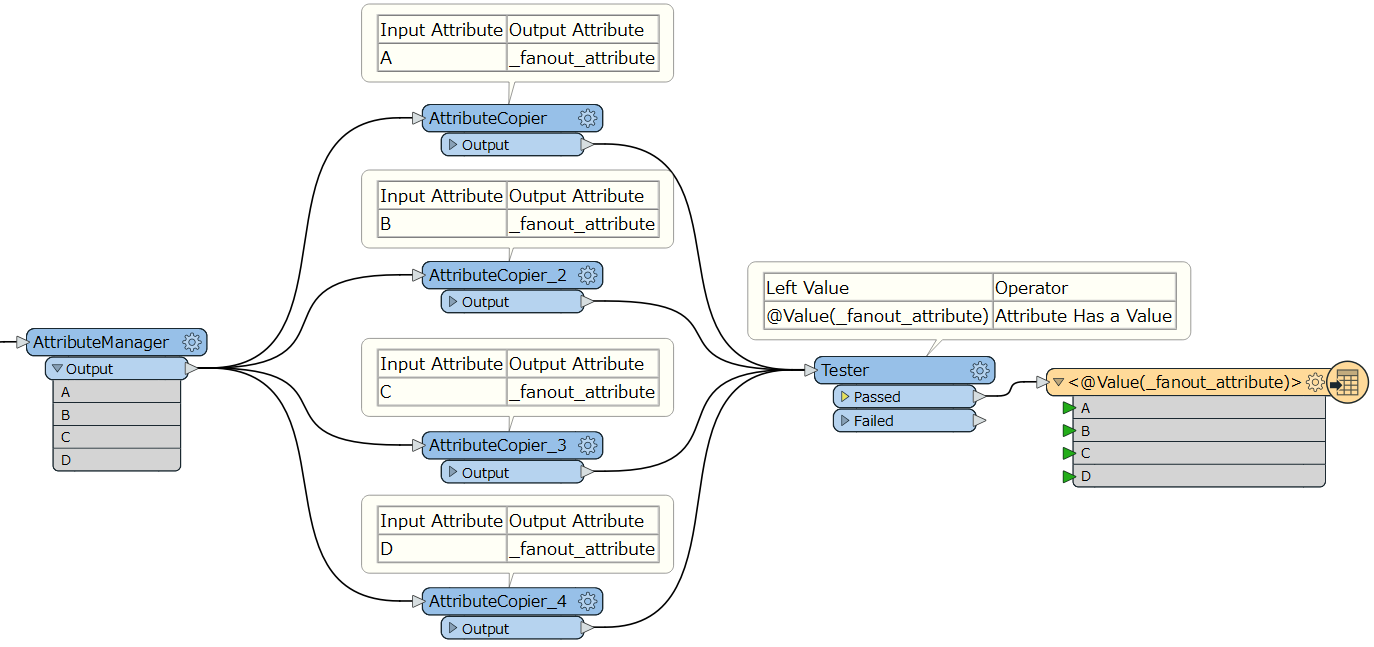
I have a dataset where want to write separate files based on multiple attributes. From the example below I would like to write a file with all the Jones, another with all the Smith, a third with all the Farmer and a fourth with any Kim. Fanout does not handle multiple attributes and in this case the StringConcatenator trick wont help, since most of the rows would be contributing to separate files.
Any ideas on how I can achieve this?
A
B
C
D
Jones
Farmer
Kim
Jones
Smith
Jones
Smith
Farmer