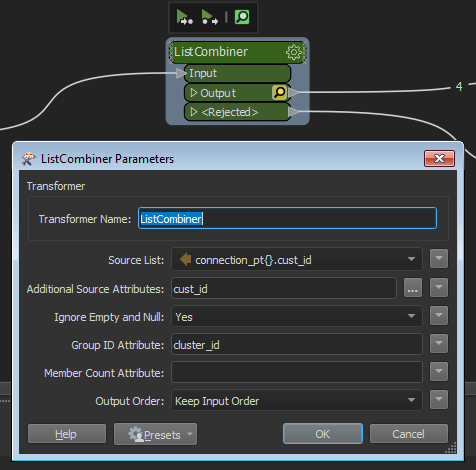
I am looking to merge into a list selected attributes from nearby points and output a single feature for each group of nearby points. PointOnPointOverlayer perfectly finds the nearby points and generates the list I want, but outputs all the input features with a list of all the other features. Thus for say 4 nearby features it will output 4 features each with a list which identifies the other 3 features in the group, but without anything like a group ID with which you could select just the first in each group with a DuplicateFilter.
This is almost exactly the example given in the PointOnPointOverlayer manual page which creates lists of crimes at common locations... but doesn't explain how to not have multiple copies of the same list at the same location. Am I missing something obvious? It seems that PointOnPointOverlayer has done all the hard work!
The closest I could get to generating a group ID is to give all my input points a unique ID field, then set "Merge attributes" to "only use incoming". This will then produce a common ID in the output for... 3 of the 4 in my group of 4, or 11 out of 12 in a group of 12.
Any suggestions? One important caveat is that the single output feature for each group can have the geometry of any one of their coordinates (but not some other location, e.g. not a centroid).