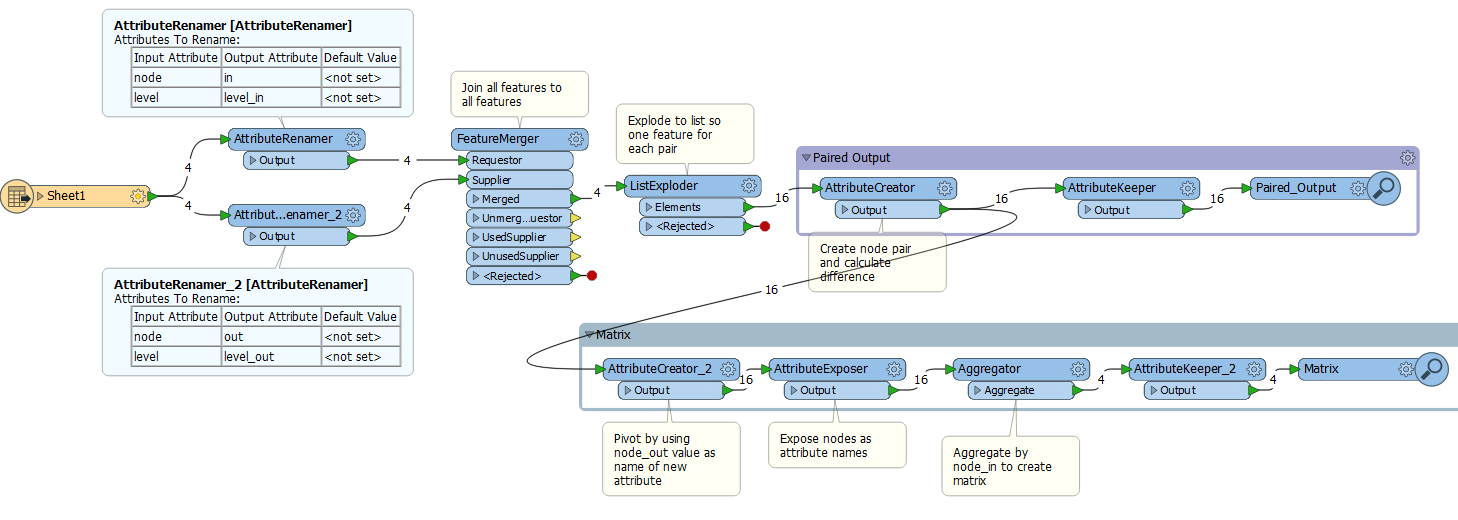
I want to take a small data set like the one below and calculate the difference between each feature as either a matrix or as a 1:1 readout
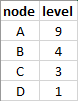
Input
Matrix output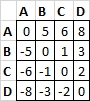
Paired output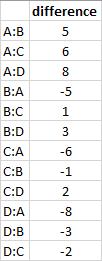
the goal would be then to use this data to evaluate a matrix of points to determine their proximity in relationship of the nodes for the given point