FME's Python interface for feature attribute manipulation seems to be mainly oriented towards small datasets, as attributes are always only accessed field by field and feature by feature. Python has a very large ecosystem around data science and data processing of very large tables, so it's often practical to load data in a dataframe and run all your computations on the dataset as a whole, to benefit from various performance optimizations around vectorization, not to mention use of fammiliar tools for people in the data science field.
Right now, loading features into a dataframe (I'm talking hundreds of columns and hundreds of thousands of rows, millions of cells) is both slow and very error-prone for a few reasons:
- Loss of schema information: getAttributeType() in the Python API can't return the full set of FME feature types, so dynamic workbenches can't properly handle things like dates unless a lot of work is done around schema detection and handling.
- Difficult to parse timestamp format: Python's standard library and most dataframe libraries can only parse fractionnal seconds up to 6 decimals, while FME does up to 9 and DatetimeNow() notably provides 7 decimals. This is a difficult problem on its own, but time series are common in the data science field, so it's especially noticeable here.
- Attribute access is per-row and field-by-field (slow): With no way to do bulk access on features, the data is accessed and converted value by value (save for lists, which can be accessed as a whole). If you have several million fields to read, that's a lot of slow python code running in very tight loops.
- Null and missing values are distinct and need to be checked separately: getAttribute() only returns None on missing values. When encountering a null value, it returns an empty string, which can throw off dataframe libraries and need to be checked for each value suceptible of returning null, adding to the overhead.
- Feature output is also per row and per feature: It's also very slow, and I belive it also blocks on waiting for downstream.
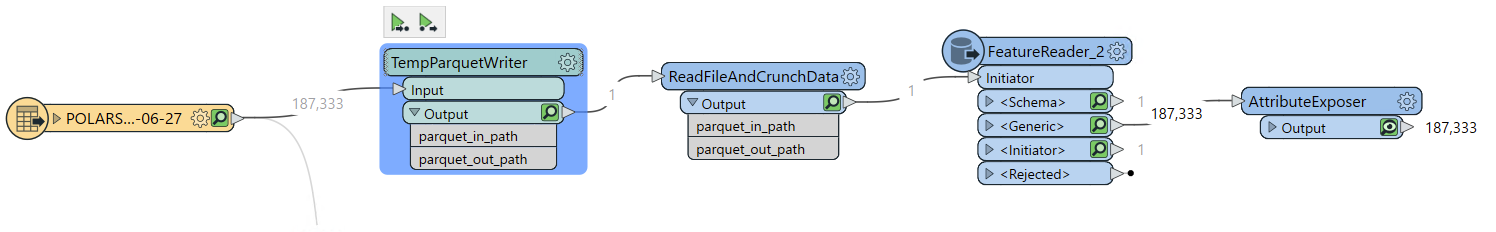
- Using files to pass data between nodes is a lot to ask: You can use feature readers and writers to create temporary files so that Python could load and dump these features in one shot, but even if I could get it to work (I haven't), it wouldn't be worth the dozen of extra nodes needed to make this work and all the visual clutter that creates.
The easy way this could be resolved, in my eyes, is to simply provide a new Python transformer (the titular DataframeTransformer) that already does the conversion of features to and from dataframes and only provides the user's code with said dataframe.
There have been efforts lately to create a standard dataframe interchange protocol throughout the Python ecosystem to encourage interoperability across libraries and avoid locking-in users into using a specific library, which Pandas supports converting to and reading from. To avoid depending on Pandas and/or its APIs, fmeobjects could expose some hypothetical "FeatureDataframe" object and leave it up to the users to load it into their dataframe library of choice.
Related questions