I have a pointcloud.las and I would like to apply some R functions on it using the lidR package. I'm having some hard time figuring out on how the pointcloud should be formatted, or passed on to the data.frame within the R Caller?
Same goes for grabbing the output.
Any clues are much appreciated
Best answer by samatsafe
View original

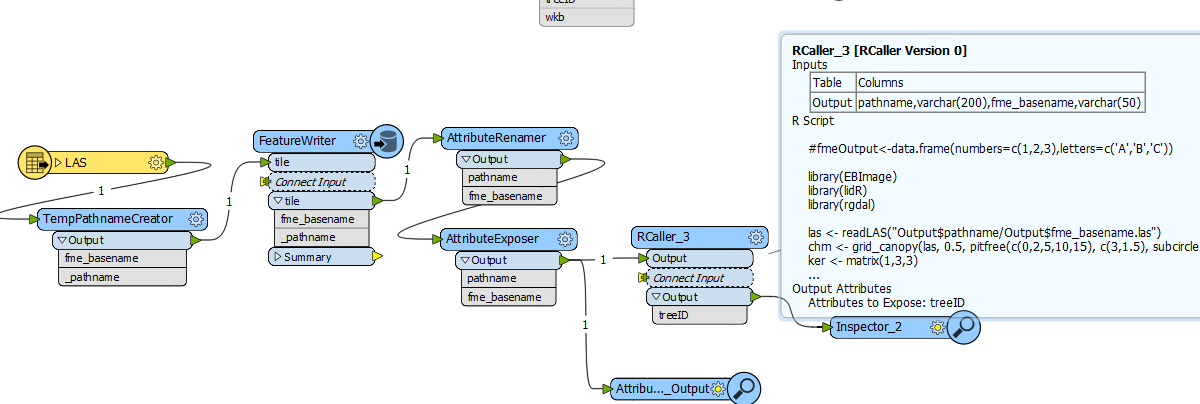 This is what my workflow looks like so far,
This is what my workflow looks like so far,