Hi All
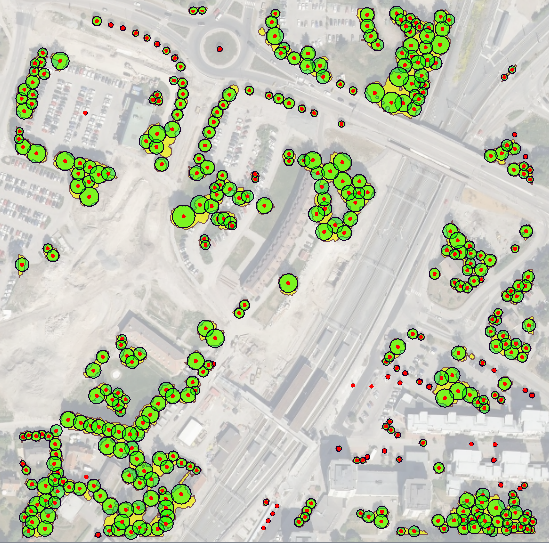
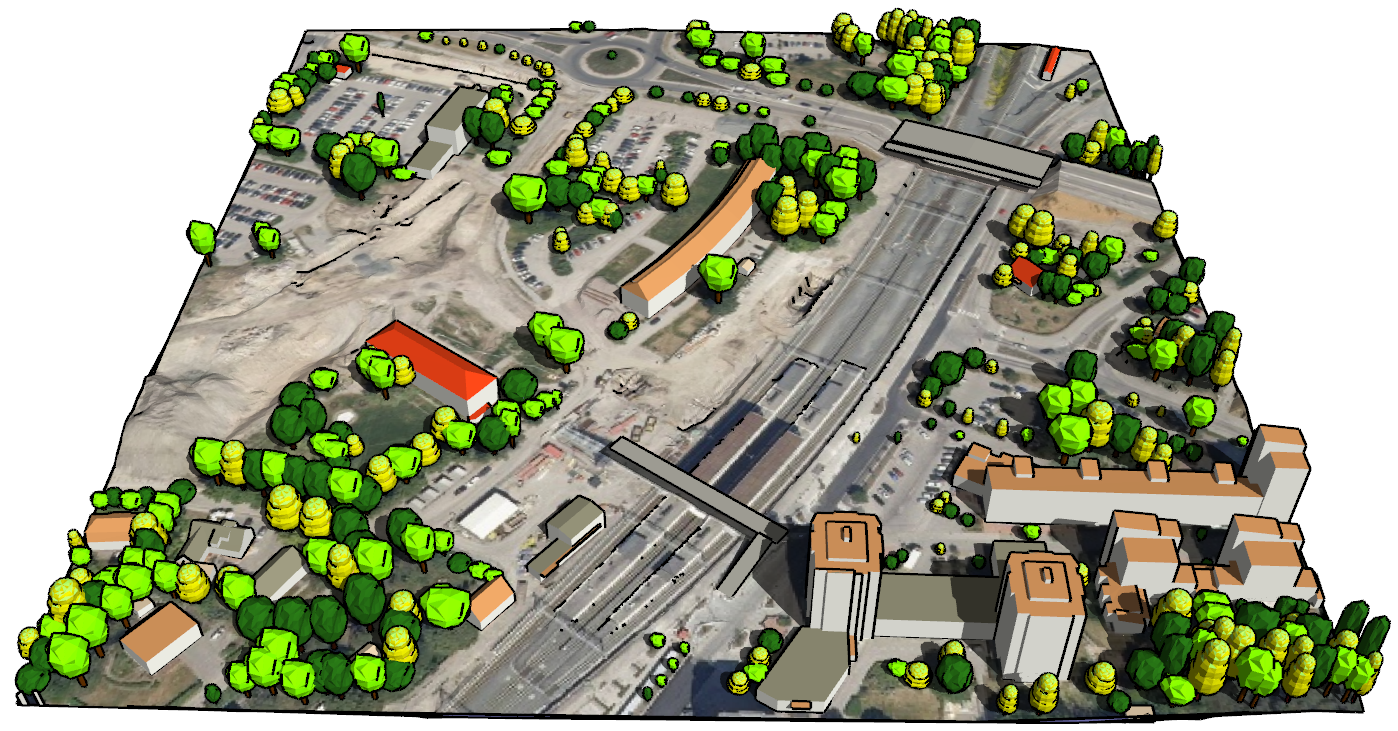
I Have the following datasets, DTM, DSM and Colour Infared, and am trying to extract trees over 3 metres in height from the data.
I know that i can subtract the DTM from the DSM to give me the height difference but can not see how to have only the raster cells over 3m, nor can i see how to extract the polygons from the imagery.
If anyone has any suggestions on how to do this then please comment.