Hi Friends,
two questions :
1. We can type the attribute names in this text field, but we cannot pass it like variable or text string(list). How can i pass this published parameter to "Attributes to remove" field to the writer dialog box?
2. If i write to shape and it crosses 2gb size fme gives error. how vcan i split the shape file if it crosses 2gb? (like fme splits v7 dgn file size in V7 writer)
Explanation :
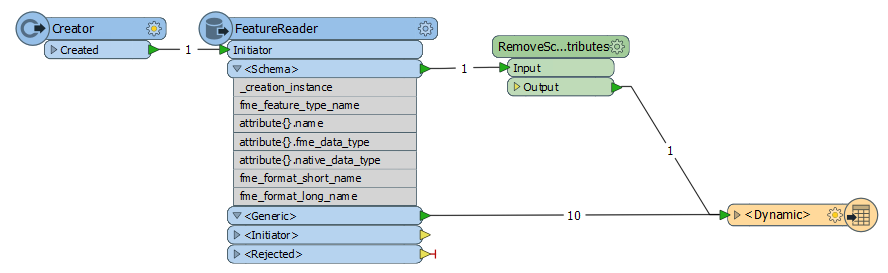
My fmw has Dynamic reader and writers. I am reading from oracle spatial (every time input tablename changes, user types in table name through published parameter) and writing to any 4 formats.(shp, tab,dgn and acad) here my fmw is dynamic. there are some attribute names that i dont want to write to destination. user types in these attribute names(space delimited). through published parameters. Every time this list changes and different attribute names comes in. In the Dynamic properties, there is a provision called "Attributes to remove". Now I have two questions :1. We can type the attribute names in this text field, but we cannot pass it like variable or text string(list). How can i pass this published parameter to "Attributes to remove" field to the writer dialog box? 2. If i write to shape and it crosses 2gb size fme gives error. how vcan i split the shape file if it crosses 2gb? (like fme splits v7 dgn file size in V7 writer)